Polyploidbreeding

PRIN 2022 (Settore LS2)
Start date: 28 September 2023
End date: 27 February 2026

Polyploidbreeding 4.0
Expanding the toolbox for cereal breeding: high-throughput genomics, 2D-3D phenomics and artificial intelligence for breeding with increasing genome complexity, from barley to durum and bread wheat (POLYPLOIDBREEDING 4.0)
Growing world population, increasing demand for high-quality protein by a rising global middle class, challenges related to climate change, and geopolitical instability are placing pressure on food production on local and global scales. At the same time, a number of major scientific and technological innovations have emerged with revolutionary impacts on agriculture, like automation (e.g. phenomics and precision farming), biotechnology (e.g. high-throughput sequencing), artificial intelligence (machine and deep learning methods). In the field of plant breeding, these scientific advancements translate to increasingly more sophisticated and technology-driven breeding methods for the genetic improvement of crops (breeding 4.0). Such efforts are typically led by diploid species, with polyploid species lagging behind. POLYPLOIDBREEDING 4.0 will focus on key technologies in diploid (barley, Hordeum vulgare) and polyploid (durum and bread wheat, Triticum durum, T. aestivum) crop species: high-throughput phenotyping (e.g. drone phenotyping, root scans from rhizotrons) and genotyping (e.g. SNP array, exome sequencing, genotyping-by-sequencing) for artificial intelligence based breeding. The objective is to develop and apply new technology-based methods for the benefit of crop breeding; barley, being a diploid species, is taken as starting point to then move to increasing levels of complexity with durum (tetraploid) and bread (hexaploid) wheat.
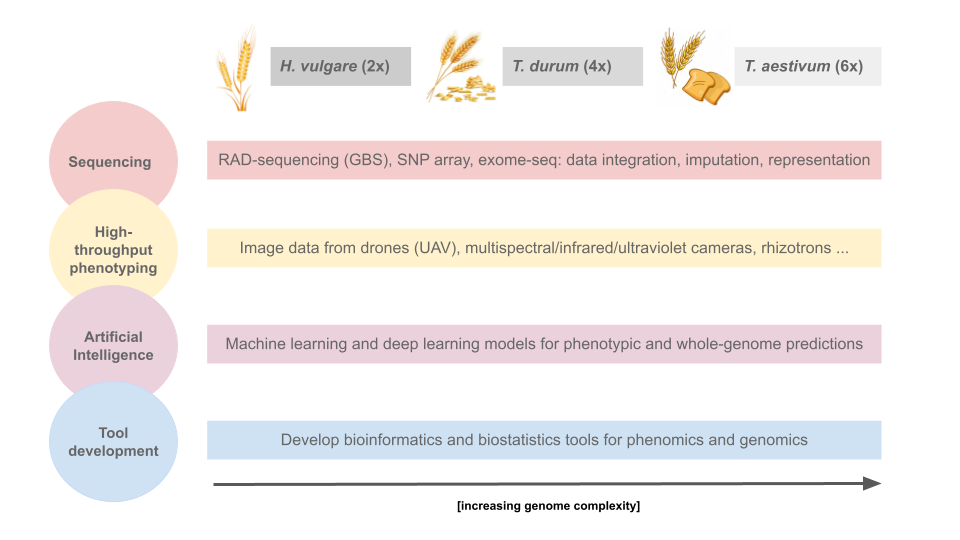
Target data are high-throughput phenotypes and genotypes, plus related environmental data (‘enviromics’, Resende et al. 2020) for
multiomics (genomics + phenomics + enviromics) predictions. Phenotypes will include yield data, morphometric measurements,
UAV (unmanned aerial vehicle: drone)-captured image data linked to morphology and production efficiency and rhizotron-based root scans (both 2D and 3D).
Multispectral cameras will capture infrared and ultraviolet wavelengths.
Machine-learning methods, focussing especially on deep learning methods, will be used for phenomic and whole-genome predictions of target phenotypes, with
innovative data representations and neural network architectures.
The project is going to unfold over two years, with these specific objectives 1) data generation: new data to fill the gaps (UAV- and
rhizotron-captured phenotypes, sequence data) and integrate new and existing/historical data; 2) modeling and tool development for
data integration, for the acquisition and processing of high-throughput phenomics and for AI-based genomic predictions, in diploid
and polyploid species; 3) calibration and fine-tuning of the main results through ad-hoc experiments (mainly in-silico) to expand the
toolbox available for cereal breeding.
Project structure
The roles and activities of each research unit in POLYPLOIDBREEDING 4.0 are schematically outlined below, organized into work packages (WP) including different Tasks (T). Beginning and end months are indicated. The project partners CNR-IBBA and UNIBO will coordinate WPs and Tasks as specified below. The subunit CREA will be involved in data generation and in deep learning modelling. The breeding company SIS will serve as external in-kind contributor and will provide data and plant material. Rhizotron phenotyping will be performed at the Forschungszentrum Jülich (Germany) in the framework of a scientific collaboration. External services and providers will be used for UAV-phenotyping and for the genotyping of new plant material.
WP 1: Project and data management (WP leader CNR-IBBA), M1-24
- T1.1: Project coordination (project management, kick-off meeting, six-month project meetings (both online and in presence), monitoring of project progress and results, monitoring partner interactions, adopting corrective measures when needed). [1-24]
- T1.2: Collection, management, and storage of all the data generated in WP2 (M3-12)
- T1.3: Integration of existing phenotypic and genotypic data (from previous projects) with newly generated data (WP2), to produce larger combined datasets to be used for phenotypic and whole-genome predictions (M10-14)
- T1.4: prepare and deposit relevant data in public repositories (e.g. the European Nucleotide Archive, ENA) (M22-24)
WP 2: Genotyping and phenotyping of plant material (WP leader UNIBO), M1-12
- T2.1: Identification of specific plant material to be genotyped and drone- and rhizotron-phenotyped among those from existing breeding programs and provided by Research Institutes , international research programmes and breeding companies. (M1-3)
- T2.2: Genotyping: identify the best technology mix for genotyping among RAD-sequencing/GBS (genotyping-by-sequencing), exome sequencing and SNP array, in view also of integration of novel and historical data. Organization of samples and genotyping strategy (M4-12)
- T 2.3: Phenotyping: organise the UAV (drone)-phenotyping and rhizotron-phenotyping sessions: frequency of UAV flights, mix of different types of cameras (visible, infrared, ultraviolet), mix of 2D and 3D root scans, subexperiments (UAV flight heights, mosaic picture vs image sampling, length of rhizotron runs etc.) (M4-12)
WP 3: Data preprocessing and tool development (WP leader CNR-IBBA), M 6-14
- T3.1: preprocessing of genotyping data: variant calling (depending on technology), quality filtering, imputation of missing genotypes. Specific methods and tools might be needed as a function of ploidy (2x, 4x, 6x) (M6-14)
- T3.2: preprocessing of drone and rhizotron phenotypes (2D and 3D image data, multiple channels, different types of cameras). Methods and tools for data acquisition, cleaning and formatting (M10-14)
- T3.3: extract traits from drone and rhizotron image/scans data through machine-learning algorithms (e.g. image segmentation, automatic labeling) and hand labeling: weed infestation, plant habit, evapotranspiration, UV light absorption, root architecture, root uptake potential and possibly others to be determined with the help of breeding companies (M10-14)
WP 4: Deep learning models for phenotypic and whole-genome predictions (WP leader CNR-IBBA), M 10-22
- T4.1: UAV assessment: develop deep learning models to predict traditionally-phenotyped traits and newly extracted traits from UAV images (WP2-3), measure model error, estimate economic impact. Evaluate impact of UAV-flight height (M10-14)
- T4.2: Rhizotron assessment: develop deep learning models to predict traits extracted from root scans (WP2-3). Develop deep learning forecasting models to predict root development based on early-stage data (M10-14)
- T4.3 Develop deep learning models for whole-genome predictions of both traditionally-phenotyped and UAV/rhizotron-extracted traits, based on genotyping data. Innovative data representations (e.g. stacked kinship matrices, genotype segments) and neural network architectures (e.g. 2D-CNN, RNN) will be explored. Compare against standard benchmark predictive models in genomic selection (GBLUP, L1/L2-penalised regression, ‘Bayesian alphabet’, RKHS). Assess model errors and estimate the economic impact in the scenario of drone- and rhizotron-based phenotyping (M10-22)
WP 5: In-silico validation and tuning of developed models and tools (WP leader UNIBO), M 16-24
- T5.1: UAV validation: develop deep learning models to predict whole land plot mosaic pictures from subsamples of few individual UAV-taken images (tools to increase the efficiency of UAV phenotyping) (M16-20)
- T5.2: Rhizotron validation: develop deep learning forecasting models to predict root development based on early-stage data (longitudinal subsampling) (M18-22).
- T5.3: Effect of ploidy: repeat the SNP calling procedure for each species using a common, fixed number of reads. Investigate the effect on predictive abilities for the available traits (M18-24) MUR - BANDO 2022Ministero dell’Università e della Ricerca
- T5.4: Environmental conditions: assess the performance of deep learning predictive models across environments (sites, years) (M18-24)
WP 6: Dissemination and public engagement (WP leader, UNIBO), M 1-36
- T6.1: Creation, maintenance and curation of project website and social media contents. (M1-24)
- T6.2: Dissemination of scientific results in publications and conferences (M8-24)
- T6.3: Fostering scientific project initiatives at the international level (M12-24)
- T6.4: Off-site and on-site training initiatives for method transfer, for project partners and open to external participants (M6-24)
References
- Resende RT et al. Enviromics in breeding: applications and perspectives on envirotypic-assisted selection. Theoretical and Applied Genetics. 2021 Jan;134(1):95-112.